Segmentation tutorials
GUI
Watch this video tutorial:
Or follow the steps below:
After opening the iCVMapper GUI, click “ICV” under the “Segmentation” tab. Wait for a new pop-up window to appear. The window should look like the image below.
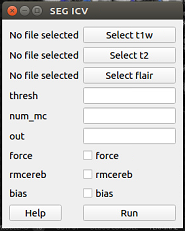
Click “Select t1w” and choose your T1 image. Click either of “Select t2w” and “Select flair” (with respect to the sequences you have) and select the correspending images. Click “Run”. Type your desired output name in the “out” box. If you want to overwrite the previous segmentation results, check the box beside “force” Check off “rmcereb” if you want to generate a segmentation with a removed cerebellum, and “bias” if you want to bias field correct the sequences using N4 Your output file will automatically appear in your t1w folder.
Command Line
icvmapper seg_icv
Optional arguments:
-h, --help show this help message and exit
-s , --subj input subject
-fl , --flair input Flair
-t1 , --t1w input T1-weighted
-t2 , --t2w input T2-weighted
-o , --out output prediction
-b, --bias bias field correct image before segmentation
-rc , --rmcereb remove cerebellum
-ign_ort, --ign_ort ignore orientation if tag is wrong
-f, --force overwrite existing segmentation
-n , --num_mc number of Monte Carlo Dropout samples
-th , --thresh threshold
-ss , --session input session for longitudinal studies
Examples:
icvmapper seg_icv -s subjectname -b
icvmapper seg_icv -t1 subject_T1_nu.nii.gz -o subject_icv.nii.gz
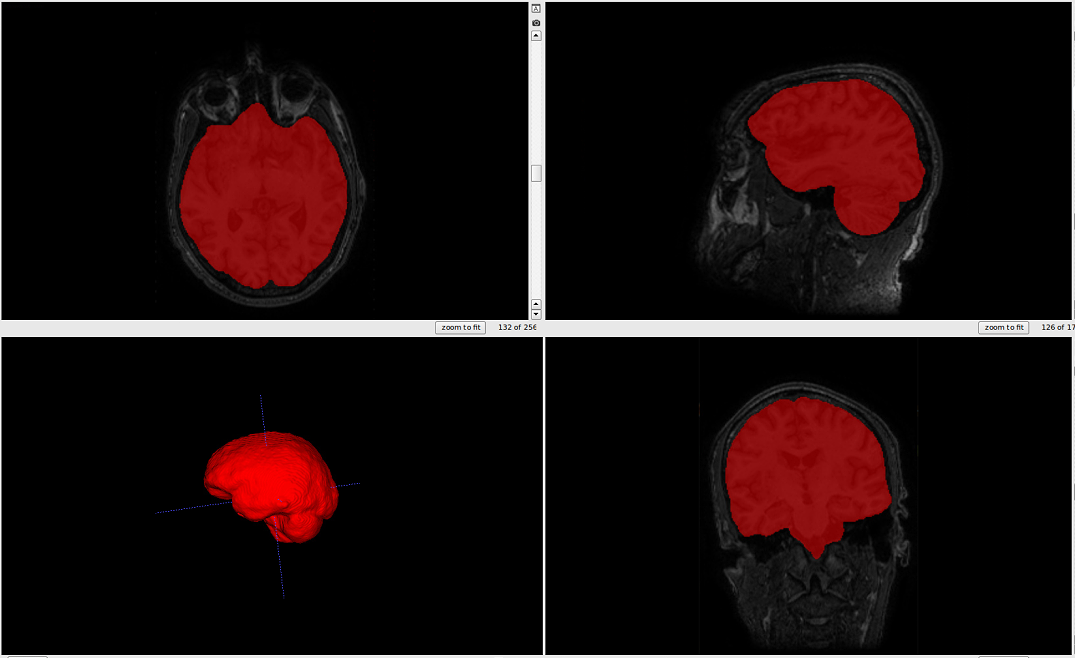
The output should look like this.: